:::
副研究員(101年迄今);助研究員(92-101年);中央研究院基因體研究中心合聘助研究員(93年迄今);國立台灣大學生化科技系合聘助理教授(98年迄今);國立台灣大學學士(78年);美國加州大學柏克萊分校碩士(82年);美國史丹佛大學博士(90年);美國史丹佛大學博士後研究(90-92年) ;
研究領域
我們實驗室擅長使用先進生物造影術擷取蛋白質巨分子的單分子結構和動態。這樣的結構資訊(通常是多重結構)代表蛋白質機器在執行功能的結構,具有生理意義。具體來說我們使用原子解析度冷凍電子顯微鏡和單分子螢光顯微術以及發展機器學習運算法術,廣泛探討控制人類遺傳訊息傳遞和修復的蛋白質巨分子,病毒的入侵,以及甲烷如何變成甲醇的機轉。目前我們破解的結構有:核醣核酸聚合酶的原子結構,魚類病毒的原子結構,還有嗜甲烷菌的膜蛋白的冷凍電子顯微鏡結構。解析和侵入細胞的機理。我們借助了酵母菌分子生物學來開發蛋白質純化和化學標記的方法,使得我們直接觀察到第二型和第三型核醣核酸聚合酶的亞單元體的振動。我們將要把這些方法應用到針對新冠肺炎核醣核酸聚合酶的藥物開發上。


重要獎項及殊榮
- 李氏基金會 (2004)
代表作
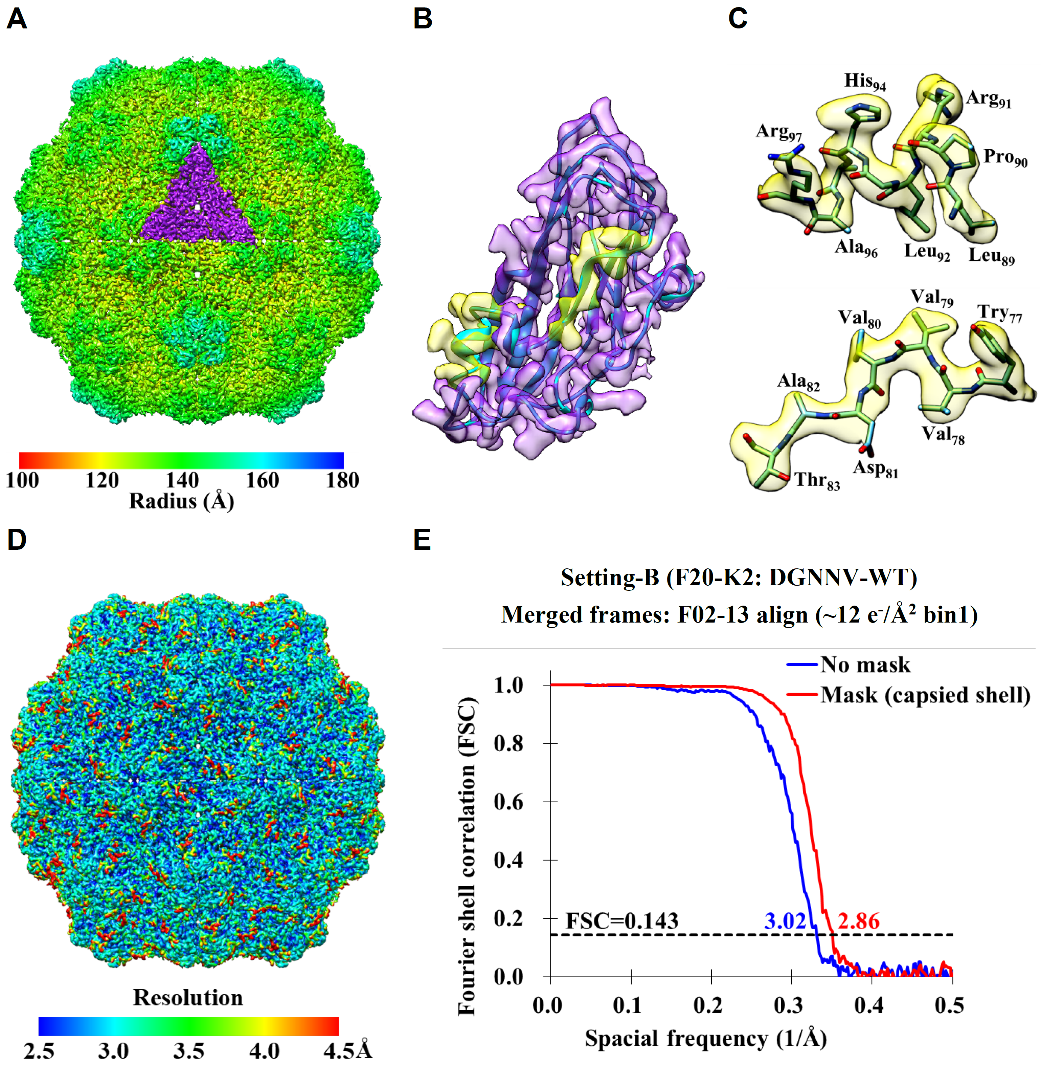
- Chang, W.-H.; Tsai, I-K.; Huang, S.-H.; Lin, H.-H.; Chung, S.-C.; Tu, I-P.; Yu, S. S.-F.; Chan, S. I.* Cryo-EM structures of the functional particulate methane monooxygenase (pMMO) from Methylococcus capsulatus (Bath) reveals the sites of the copper centers. Journal of American Chemical Society 2021, In Press.
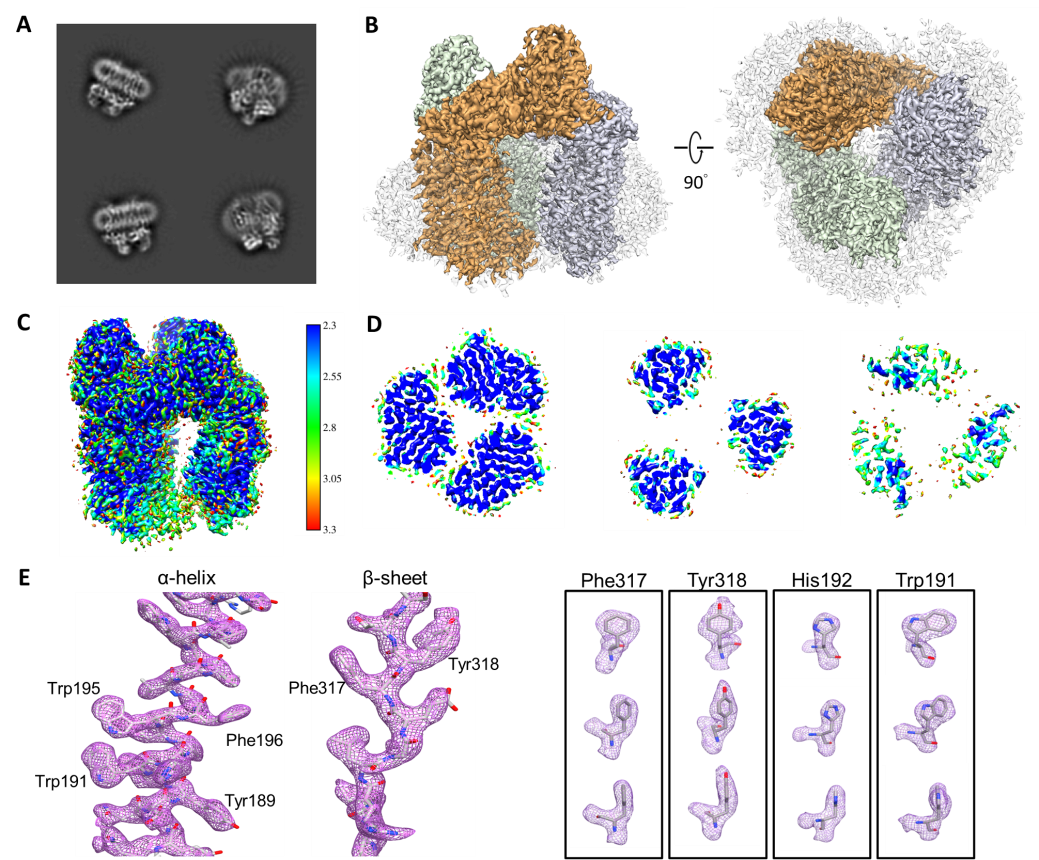
- Wang, C.-H.; Chen, D.-H.; Huang, S.-H.; Wu, Y.-M.; Chen, Y.-Y.; Hwu, Y.; Bushnell, D.; Kornberg, R.; Chang, W.-H. Sub-3 Å cryo-EM structures of necrosis virus particles via the use of multi-purposed TEM with electron counting camera, Intl J. of Mol. Science 2021, Accepted.
- Yeh, F.-L.; Chang, S.-L.; Ahmed, G. R.; Tung, L.; Lanier, L. S.; Maeder, C.; Lin, C.-M.; Tsai, S.-C.; Chang, W.-H.; Chang, T.-H. (2021) Activation of Prp28 ATPase by phosphorylated Npl3 at a critical step of spliceosome remodeling. Nature Commun. 2021, 12, Article number: 3082
- Chung, S.-c.; Lin, H.-H.; Niu, B.-Y.; Huang, S.-H.; Tu, I-P.; Chang, W.-H. Pre-Pro is a fast pre-processor for single particle cryo-EM by enhancing 2D classification. Commun. Biol. 2020, 3, Article number: 508.
- Huang, C.-F.; Chang, W.-H.; Lee, T.-K.; Joti, Y.; Nishino, Y.; Kimura, T.; Suzuki, A.; Bessho, Y.; Lee, T.-T.; Chen, M.-C.; Yang, S.-M.; Hwu, Y.; Huang, S.-H.; Li, P.-N.; Chen, P.; Tseng, Y.-C.; Ma, C.; Hsu, T.-L.; Wong, C.-H.; Tono, K.; Ishikawa, T.; Liang, K. S. XFEL coherent diffraction imaging for weakly scattering particles using heterodyne interference. AIP Adv. 2020, 10 (5), 055219.
- Štěrbová, P.; Wu, D.; Lou, Y.-C.; Wang, C.-H.; Chang, W.-H.; Tzou, D.-L. M. NMR assignments of protrusion domain of capsid protein from dragon grouper nervous necrosis virus. Biomol NMR Assign 2019, 14, 63-66.
- Wu, J.-S.; Chen, T.-Y.; Lin, S. S.-Y.; Lin, S.-Y.; Hung, C.-Y.; Tu, I-P.; Chen, H.-T.; Chang, W.-H. Deriving a sub-nanomolar affinity peptide from TAP to enable smFRET analysis of RNA polymerase II complexes. Methods 2019, 159-160, 59-69.
- Wu, Y.-M.; Wang, C.-H.; Chen, Y.-Y.; Miyazaki, N.; Murata, K.; Nagayama, K.; Chang, W.-H. Zernike phase contrast cryo-electron microscopy reveals 100 kDa component in a protein complex, J. Phys. D: Appl. Phys. 2013, 46, 494008 (special issue on phase contrast)
- Chang, J.-W.; Wu, Y.-M.; Chen, Z.-Y.; Huang, S.-H.; Wang, C.-H.; Wu, P.-l.; Weng, Y.-P.; You, C.; Piehler, J.; Chang, W.-H. Hybrid Electron Microscopy-FRET imaging localizes the dynamical C-terminus of Tfg2 in RNA polymerase II-TFIIF with nanometer precision. J. Struct. Biol. 2013, 184, 52-62 (special issue on hybrid methods)
- Wu, Y.-M.; Chang, J.-W.; Wang, C.-H.; Lin, Y.-C.;Wu, P.-L.; Huang, S.-H.; Chang, C.-C.; Hu, X.; Gnatt, A.; Chang, W.-H. Regulation of mammalian transcription by Gdown1 through a novel steric crosstalk revealed by cryo-electron microscopy, EMBO J. 2012, 31, 3575-3587
- Chang, W.-H.; Chiu, M. T.-K.; Chen, C.-Y.; Yen, C.-F.; Lin, Y.-C.; Cheng, H.; Fu, J.; Tu, I-P. Zernike Phase plate cryo-electron microscopy facilitates single particle analysis of asymmetric proteins, Structure 2010, 18, 17-27
- Chen, C.-Y.; Chang, C.-C.; Yen, C.-F.; Chiu, M. T.-K.; Chang, W.-H. Mapping RNA exit on RNA polymerase II by FRET analysis, PNAS 2009, 106, 127-132
Update: 2021-06-29
期刊論文
- Štěrbová Petra, Wang Chun-Hsiung, Carillo Kathleen J.D., Lou Yuan-Chao, Kato Takayuki, Namba Keiichi, Tzou Der-Lii M., Chang Wei-Hau Cryo-EM inspired NMR analysis reveals a pH-induced conformational switching mechanism for imparting dynamics to Betanodavirus protrusions. BioRxiv 2024-03, .
- Lin Hsin-Hung, Wang Chun-Hsiung, Wu Yi-Min, Huang Shih-Hsin, Lin Sam Song-Yao, Kato Takayuki, Namba Keiichi, Hosogi Naoki, Song Chihong, Murata Kazuyoshi, Yen Ching-Hsuan, Hsu Tsui-Ling, Wong Chi-Huey, Tu I-Ping, Chang Wei-Hau Use of phase plate cryo-EM reveals conformation diversity of therapeutic IgG with 50 kDa Fab fragment resolved below 6 Å. BioRxiv 2023-12-22, .
- Shekhar AC, Sun YE, Khoo SK, Lin YC, Malau EB, 「Chang WH」, Chen HT* Site-directed biochemical analyses reveal that the switchable C-terminus of Rpc31 contributes to RNA polymerase III transcription initiation.. Nucleic acids research 2023-05, 51, 4223-4236.
- Lai Tze Leung, Wang Shao-Hsuan, Chung Szu-Chi, Chang Wei-hau, Tu I-Ping Uncertainty Quantification in Dynamic Image Reconstruction with Applications to Cryo-EM. Statistica Sinica 2023, 33, 1771-1788.
- 「Wei-Hau Chang」*, Shih-Hsin Huang,, Hsin-Hung Lin, Szu-Chi Chung, I-Ping Tu Cryo-EM Analyses Permit Visualization of Structural Polymorphism of Biological Macromolecules.. Frontiers in Bioinformatics 2021-12, 1, 788308.
- Sunney I. Chan*, 「Wei-Hau Chang」*, Shih-Hsin Huang, Hsin-Hung Lin, Steve Sheng-Fa Yu* Catalytic Machinery of Methane Oxidation in Particulate Methane Monooxygenase (pMMO). Journal of Inorganic Biochemistry 2021-09, 225, 111602.
- Chun-Hsiung Wang, Dong-Hua Chen, Shih-Hsin Huang , Yi-Min Wu, Yi-Yun Chen, Yeukuang Hwu, David Bushnell, Roger Kornberg, 「Wei-Hau Chang」* Sub-3 Å Cryo-EM Structures of Necrosis Virus Particles via the Use of Multipurpose TEM with Electron Counting Camera. International Journal of Molecular Sciences 2021-06, 22(13), 6859.
- Fu-Lung Yeh, Shang-Lin Chang, Golam Rizvee Ahmed, Hsin-I Liu, Luh Tung, Chung-Shu Yeh, Leah Stands Lanier, Corina Maeder, Che-Min Lin, Shu-Chun Tsai, Wan-Yi Hsiao, 「Wei-Hau Chnag」, Tien-Hsien Chang* Activation of Prp28 ATPase by phosphorylated Npl3 at a critical step of spliceosome remodeling. Nature Communications 2021-05, 12(1), 3082.
- Wei-Hau Chang*, Hsin-Hung Lin, I-Kuen Tsai, Shih-Hsin Huang, Szu-Chi Chung, I-Ping Tu, Steve S.-F. Yu* and Sunney I Chan* Copper Centers in the Cryo-EM Structure of Particulate Methane Monooxygenase Reveal the Catalytic Machinery of Methane Oxidation. Journal of the American Chemical Society 2021, 143(26), 9922-9932.
- Wang Shao-Hsuan, Yao Yi-Ching, Chang Wei-Hau, Tu I-Ping* Quantification of model bias underlying the phenomenon of Einstein from Noise. Statistica Sinica 2020-11, .
- Szu-chi Chung, Shao-hsuan Wang, Bo-yao Niu, Su-yun Huang, Wei-hau Chang, I-Ping Tu* Two-stage dimension reduction (2SDR) for noisy high dimensional images and application to cryogenic electron microscopy. Annals of Mathematical Sciences and Applications 2020-10, 5(2), 283-316.
- Szu-chi Chung, Hsin-Hung Lin, Bo-Yao Niu, Shih-Hsin Huang, I-Ping Tu*, 「Wei-hau Chang」* Pre-Pro is a fast pre-processor for single particle cryo-EM by enhancing 2D classification. Communications Biology 2020-09, 3, 508.
- Chi-Feng Huang, Wei-Hau Chang*, Ting-Kuo Lee, Yasumasa Joti, Yoshinori Nishino, Takashi Kimura, Akihiro Suzuki, Yoshitaka Bessho, Tsung-Tse Lee, Mei-Chun Chen, Shun-Min Yang, Yeukuang Hwu, Shih-Hsin Huang, Po-Nan Li, Peilin Chen, Yung-Chieh Tseng, Che Ma, Tsui-Ling Hsu, Chi-Huey Wong, Kensuke Tono, Tetsuya Ishikawa, Keng S. Liang. XFEL coherent diffraction imaging for weakly scattering particles using heterodyne interference. AIP Advances 2020-05, 10(5), 055219.
- Petra Štěrbová, Danni Wu, Yuan-Chao Lou, Chun-Hsiung Wang, Wei-Hau Chang*, Der-Lii M Tzou*. NMR assignments of protrusion domain of capsid protein from dragon grouper nervous necrosis virus. Biomolecular NMR Assignments 2020-04, 14(1), 63-66.
- Jheng-Syong Wu, Tzu-Yun Chen, Sam Song-Yao Lin, Shu-Yu Lin, Cheng-Yu Hung, I-Ping Tu, Hung-Ta Chen, Wei-Hau Chang* Deriving a sub-nanomolar affinity peptide from TAP to enable smFRET analysis of RNA polymerase II complexes. METHODS 2019-02, pii: S1046-2023(18)30301-3. doi: 10.1016/j.ymeth.2.
- Huang Chi-Feng, Liang Keng S., Hsu Tsui-Ling, Lee Tsung-Tse, Chen Yi-Yun, Yang Shun-Min, Chen Hsiang-Hsin, Huang Shih-Hsin, Chang Wei-Hau, Lee Ting-Kuo, Chen Peilin, Peng Kuei-En, Chen Chien-Chun, Shi Cheng-Zhi, Hu Yu-Fang, Margaritondo Giorgio, Ishikawa Tetsuya, Wong Chi-Huey, Hwu Y. Free-electron-laser coherent diffraction images of individual drug-carrying liposome particles in solution. Nanoscale 2018, 10(6), 2820-2824.
- Min-Yuan Chia, Wan-Yu Chung, Chung-Hsiung Wang, Wei-Hau Chang, Min-Shi Lee* Development of A High-Growth Enterovirus 71 Vaccine Candidate Inducing Cross-Reactive Neutralizing Antibody Responses. VACCINE 2018, 21;36(9):1167-1173.
- Wong HC, Wang TY, Yang CW, Tang CT, Ying C, Wang CH, Chang WH Characterization of a lytic vibriophage VP06 of Vibrio parahaemolyticus.. Research in microbiology 2018, pii: S0923-2508(18)30117-7.
- Chung-Shu Yeh, Shang-Lin Chang, Jui-Hui Chen, Hsuan-Kai Wang, Yue-Chang Chou, Chun-Hsiung Wang, Shih-Hsin Huang, Amy Larson, Jeffrey A Pleiss, Wei-Hau Chang, Tien-Hsien Chang* The conserved AU dinucleotide at the 5′ end of nascent U1 snRNA is optimized for the interaction with nuclear cap-binding-complex. Nucleic Acids Research 2017, 45(16), 9679-9693.
- Ting-Li Chen*, Dai-Ni Hsieh*, Hung Hung, I-Ping Tu*, Pei-Shien Wu, Yi-Ming Wu*, Wei-Hau Chang*, Su-Yun Huang* $gamma$-SUP: A clustering algorithm for cryo-electron microscopy images of asymmetric particles. The Annals of Applied Statistics 2014, 8(1), 259-285.
- Yi-min Wu, Chu-Hsiung Wang, Jen-wei Chang, Kazuyoshi Murata, Kuniaki Nagayama, Wei-hau Chang* Zernike phase contrast cryo-electron microscopy reveals 100 kDa component in a protein complex. JOURNAL OF PHYSICS D-APPLIED PHYSICS 2013-12, 46(49), 494008-494017.
- Jen-Wei Chang, Yi-Min Wu, Zi-Yun Chen, Shih-Hsin Huang, Chun-Hsiung Wang, Pei-lun Wu, Yi-ping Weng, Changjiang You, Jacob Piehler, Wei-hau Chang* Hybrid electron microscopy-FRET imaging localizes the dynamical C-terminus of Tfg2 in RNA polymerase II–TFIIF with nanometer precision. JOURNAL OF STRUCTURAL BIOLOGY 2013-07, 184(1), 52-62.
- Yi-min Wu, Jen-wei Chang, Chun-hsiung Wang,Yen-chen Lin, Pei-lun Wu, Shih-hsin Huang, Chia-chi Chang, Xiaopeng Hu, Averell Gnatt*, Wei-hau Chang * Regulation of mammalian transcription by Gdown1 through a novel steric crosstalk revealed by cryo-electron microscopy. EMBO JOURNAL 2012-08, 31(17), 3575-3587.
- Hou-chih Lee, Bo-lin Lin, Wei-hau Chang *, I-ping Tu* Toward automated de-noising of single-molecule FRET data: ADN for smFRET. JOURNAL OF BIOMEDICAL OPTICS 2012-02, 17, 011007.
- Jiahn-Haur Liao, Chiao-I Kuo, Ya-Yi Huang, Yu-Ching Lin, Yen-Chen Lin, Chen-Yui Yang, Wan-Ling Wu, Wei-Hau Chang, Yen-Chywan Liaw, Li-Hua Lin, Chung-I Chang, Shih-Hsiung Wu* A Lon-Like Protease with No ATP-Powered Unfolding Activity. PLOS ONE 2012, PLoS ONE 7(7): e40226.
- Hung-Ju Wei , Weihau Chang , Shih-Chang Lin , Wen-Chun Liu , Ding-Kao Chang , Pele Chong , Suh-Chin Wu* Fabrication of influenza virus-like particles using M2 fusion proteins for imaging single viruses and designing vaccines. Vaccine 2011-09, 29, 7163-7172.
- CC Fu, CF Lin, QZe Gao, WZ Yang, TS Lim,LL Yang, CF Yen, Wei-Hau Chang, Hanna S. Yuan d, S-Y Sheu,Dah-Yen Yang, Wunshain Fann Fis-protein induces rod-like DNA bending. Chemical Physics Letters 2010-10, 500,318-322.
- Wang CH, Hsu CH, Wu YM, Luo YC, Tu MH, Chang WH, Cheng RH, Lin CS* Roles of cysteines Cys115 and Cys201 in the assembly and thermostability of grouper betanodavirus particles. VIRUS GENES 2010-08, 41, 73-80.
- Yen-Chen Lin, Bo-lin Lin, Tommy Setiawan, Chia-Chi Cheng, Chi-Fu Yen, Wei-hau Chang* A single molecule FRET study of formation of RNA polymerase II elongation complex on passivated surface . Journal of Chinese Chemical Society 2010-07, 57(3B), 514-521.
- Wei-hau Chang, Yu-hua Liu Bioorthogonal labeling method for single molecule FRET. Journal of Chinese Chemical Society 2010-07, 57(3B),505-513.
- Wei-hau Chang, Michael T.-K. Chiu, Chin-Yu Chen, Chi-Fu Yen, Yen-Cheng Lin, Yi-ping Weng, Ji-chau Chang, Yi-min Wu, Holland Cheng, Jianhua Fu, I-ping Tu Zernike phase plate cryo-electron microscopy facilitates single particle analysis of unstained asymmetric protein complexes. Structure 2010-01, 18, 17-27.
- Velusamy M, Huang JH, Hsu YC, Chou HH, Ho KC, Wu PL, Chang WH, Lin JT, Chu CW* Dibenzo[f,h]thieno[3,4-b] quinoxaline-based small molecules for efficient bulk-heterojunction solar cells. Org. Lett. 2009-11, 11,4898-4901.
- Jiessie Shiue, Chia-Seng Chang, Sen-Hui Huang, Chih-Hao Hsu, Jin-Sheng Tsai, Wei-hau Chang, Yi-Min Wu, Yen-Chen Lin, Pai-Chia Kuo, Yan-Shan Huang, Yeukuang Hwu, Ji-Jung Lai, Fang-Gang Tseng, and Fu-Rong Chen Phase TEM for biological imaging utilizing a Boersch elecrostatic phase plate: theory and practice. Journal of Electron Microscopy 2009, 58,137-145.
- Chen CY, Chang CC, Yen CF, Chiu MT, *Chang WH Mapping RNA exit channel on transcribing RNA polymerase II by FRET analysis.. Proceedings of the National Academy of Sciences of the United States of America 2009, 106(1), 127-32.
- Yi-Min Wu, Chi-Hsin Hsu, Chun-Hsiung Wang, Wangta Liu, Wei-hau Chang* and Chan-Shing Lin* Role of DxxDxD motif in the assembly and stability of the betanodavirus particles. ARCHIVES OF VIROLOGY 2008-08, 153:1633-1642.
- Chang WH, Kornberg RD Electron crystal structure of the transcription factor and DNA repair complex, core TFIIH.. Cell 2000, 102(5), 609-13.



